User:Eaberry/sandbox
Sub- and Super-scripts[edit]
Use the HTML tags "sub" and "sup": I22
| Names | |
|---|---|
| IUPAC name
2-[2-[2-[2-(2-dodecoxyethoxy)ethoxy]ethoxy]ethoxy]ethanol
| |
| Other names
Dodecylpentaglycol
Laureth-5 | |
| Identifiers | |
3D model (JSmol)
|
|
| Abbreviations | C12E5 |
| ChEBI | |
| ChemSpider | |
PubChem CID
|
|
| |
| |
| Properties | |
| C22H46O6 | |
| Molar mass | 406.59704 |
| Density | 0.963 g/mL at 20 °C |
| Surface tension: | |
| 7×10−5 M at 25 °C.[1] | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
What is the pKa?[edit]
The text gives the pKa as 8.00 in one place, and 7.7 in another. Sigma-Aldrich reports 7.92. I suspect the iargest value is the thermodynamic pKa extrapolated to infinite dilution, and the other two are at more reasonable ionic strengths. Because the dianionic red form is stabilized by high ionic strength more than the mono-anionic yellow form, ioonic strength will favor dissociation, making it a stronger acid. Does anyone have a reference to the determination of the pKa, preferably with ionic strength dependence?
I purchased the sodium salt from sigma. When dissolved in the standard pH 7.02 phosphate buffer using for calibrating pH electrodes, the molar ratio of the dibasic 559 cpd to monobasic 432 nm compound was 0.187, consistent with a pKa of 7.75 at this ionic strength (which I suspect is rather high).
Dependence of pH on ionic strength and temperature[edit]
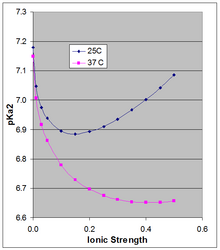
The Henderson-Hasselbach equation gives the pH of a solution relative to the pKa of the acid-base pair. However the pKa is dependent on ionic strength and temperature, and as it shifts so will the pH of a solution based on that acid-base pair. Because the doubly charged HPO4-2 is stabilized more by high ionic strength than is the singly-charged H2PO4-, their pKa is somewhat dependent on ionic strength. The thermodynamic pKa of ~7.2 is the value extrapolated to zero ionic strength, and is not applicable at physiological ionic strength.
Phillips et al.[3] measured the pKa at 10, 25, and 37°C at various ionic strength. For the latter two temperatures they report pKa in Debye-Hückel equations (plotted in the accompanying figure for µ up to 0.5 M):
at 25C: pKa2 = 7.18 - 1.52 sqrt(µ) + 1.96 µ
at 37C: pKa2 = 7.15 - 1.56 sqrt(µ) + 1.22 µ
The pKa0 is weakly dependent on temperature. Phillips et al. reported ∆H0 at 25C of 760 cal/mol (3180 J/mol) and a linear dependence of pKa0 on 1/T (Van 't Hoff equation). The positive ∆H0 results in an increase in Ka, and thus a decrease in pKa0 with rising temperature, the change in pKa0 being 166 × the change in (1/T), which around 25C results in a change in pKa0 of -0.00187 per degree. This applies strictly to the extrapolated thermodynamic pKa0 at infinite dilution, and as the Figure shows, the temperature effect can be much larger at higher ionic strength.
Latex[edit]
| Original author(s) | Leslie Lamport |
|---|---|
| Platform | Cross-platform |
| Type | Typesetting |
| License | LaTeX Project Public License (LPPL) |
| Website | www.latex-project.org |
LaTeX (formatted as LaTeX, pronounced /ˈlɑːtɛx/, /ˈlɑːtɛk/, /ˈleɪtɛx/, or /ˈleɪtɛk/), is a document markup language and document preparation system for the TeX typesetting program. The term LaTeX refers only to the language in which documents are written, not to the editor application used to write those documents. In order to create a document in LaTeX, a .tex file must be created using some form of text editor. While most text editors can be used to create a LaTeX document, a number of editors have been created specifically for working with LaTeX.
LaTeX is widely used in academia.[4][5] It is also used as the primary method of displaying formulas on Wikipedia. As a primary or intermediate format, e.g., translating DocBook and other XML-based formats to PDF, LaTeX is used because of the high quality of typesetting achievable by TeX. The typesetting system offers programmable desktop publishing features and extensive facilities for automating most aspects of typesetting and desktop publishing, including numbering and cross-referencing, tables and figures, page layout and bibliographies.
LaTeX is intended to provide a high-level language that accesses the power of TeX. LaTeX essentially comprises a collection of TeX macros and a program to process LaTeX documents. Because the TeX formatting commands are very low-level, it is usually much simpler for end-users to use LaTeX.
LaTeX was originally written in the early 1980s by Leslie Lamport at SRI International.[6] The current version is LaTeX2e (styled as LaTeX2ε). LaTeX is free software and is distributed under the LaTeX Project Public License (LPPL).{{MaTeX2e}}
Typesetting system[edit]
LaTeX is based on the philosophy that authors should be able to focus on the content of what they are writing without being distracted by its visual presentation. In preparing a LaTeX document, the author specifies the logical structure using familiar concepts such as chapter, section, table, figure, etc., and lets the LaTeX system worry about the presentation of these structures. It therefore encourages the separation of layout from content while still allowing manual typesetting adjustments where needed. This is similar to the mechanism by which many word processors allow styles to be defined globally for an entire document or the use of Cascading Style Sheets to style HTML.
Structure and Mechanism[edit]
The X-ray structures of yeast MPP, and a cleavage-deficient MPP with substrate bound, are available[7]. The sequence around the scissile bond binds as a beta strand in such a way that the scissile bond aligns with the zinc binding site to allow nucleophilic attack by a zinc--coordinated water molecule on the carbonyl carbon of the residue before the scissile bond (last residue of the signal peptide).
How to use a template[edit]
You transclude it just as was done in Thames, by using {{infobox river}} and then including whichever parameters you wish to use. You might find it easier to copy the blank from the "Usage" section from Template:Infobox river/doc and then fill in the values for each parameter you wish to use (and you might want to tidy up by deleting the parameters which you aren't using). - David Biddulph (talk) 12:13, 5
- Here
Bouchard Creek Physical characteristics Length 40 mi Width • minimum 2 m • maximum 20 m
Wiki-Math[edit]
When each receptor has a single ligand binding site, the system is described by
with an on-rate (kon) and off-rate (koff) related to the dissociation constant through Kd=koff/kon. When the system equilibrates,
so that the average number of ligands bound to each receptor is given by
which is the Scatchard equation for n=1.
n = [RL] + [R];
[R] = n - [RL] (2)
From (1):
Substituting for [R] from (2)
plotting [RL] vs [RL]/[L] (bound vs bound/free) give n as the Y intercept, -Kd as the slope.
Molecular target[edit]
Buparvaquone resistance appears to be associated with parasite mutations in the Qo quinone-binding site of mitochondrial cytochrome b [8]. Its mode of action is thus likely to be similar to that of the antimalarial drug atovaquone, a similar 2-hydroxy-1,4-napthoquinone that binds to the Qo site of cytochrome b thus inhibiting Coenzyme Q – cytochrome c reductase.
Dinoseb[edit]
Dinoseb is also a weak inhibitor of Complex III[9] and Complex II of the repiratory chain.[10]
Mitochondrial vs Cytoplasmic Protein Synthesis[edit]
Mitochondrial protein synthesis is resistant to inhibition by cycloheximide. On the other hand chloramphenicol inhibits mitochondrial (and bacterial) protein synthesis, but synthesis on mitochondrial ribosomes is resistant. Before genomes were available, this dichotomy was used to determine which mitochondrial proteins were synthesized in the cytoplasm from nuclear genes[12].
At higher concentrations, stigmatellin also inhibits Complex I, as a "Class B" inhibitor of that enzyme [13]
Crystal structures for stigmatellin-inhibited bc1 complex from bovine, avian, yeast (Saccharomyces cerevisiae) and bacterial (Rhodobacter capsulatus, Cereibacter sphaeroides, and Paracoccus denitrificans) sources are available.
The chromone inhibitor stigmatellin--binding to the ubiquinol oxidation center at the C-side of the mitochondrial membrane.
von Jagow G, Ohnishi T. FEBS Lett. 1985 Jun 17;185(2):311-5. doi: 10.1016/0014-5793(85)80929-7. PMID: 2987042 [14]
[15]
In 1979 Trumpower's lab isolated the "oxidation factor" from bovine mitochondria and showed it was a reconstitutively-active form of the Rieske Iron-sulfur protein
Chicken ovomucoid is synthesized as a pre-protein consisting of 208 or 210 amino acids. A 24-residue presequence is removed [18] to give the mature 20 kDa protein of 185 residues. Ovomucoid has serine protease inhibitor activity. The structure of turkey ovomucoid complexed with the serine protease subtilisin has been determined by X-ray crystallography. [19]
Template[20]
Furthermore, the "intensity statistics" of centric reflections
( )
<gnuplot>
set xrange [0:5]
plot 2*x*exp(-x*x)
</gnuplot>
are different from those of acentric reflections
( )
<gnuplot>
set xrange [0:5]
plot sqrt(2/3.14)*exp(-x*x/2)
</gnuplot>
J[edit]
| Ext. | Description | Used by |
|---|---|---|
| JL | Julia script file | Julia (programming language) |
| J2C[21] | JPEG 2000 image | JPEG 2000 |
| JAR[22] | Java archive | JAR, Java Games and Applications |
| JAV[23] | see JAVA | |
| JAVA[23] | Java source code file | |
| JBIG[24][25][26] | Joint Bilevel Image Group | |
| JNLP[27] | Java Network Launching Protocol | Java Web Start |
| JP2[21] | JPEG 2000 image | |
| JPEG[28] | Joint Photographic Experts Group graphics file format | Minolta/Konica Minolta cameras use this for JPEGs in Adobe RGB color space |
| JPEG[28] | Joint Photographic Experts Group graphics file format | QPeg - FullView - Display |
| JPEG[28] | Joint Photographic Group | various (Minolta/Konica Minolta cameras use this for JPEGs in sRGB color space) |
| JS[29] | JavaScript file | script in HTML pages |
| JSON[30][31] | JSON (JavaScript Object Notation) | Ajax |
| JSP[32] | Jakarta Server Pages | Dynamic pages running Web servers using Java technology |
ATPase[edit]
Further updates: These figures may still require further tweaking as new structural details become available. The above value of 3 H+/ATP for the synthase assumes the synthase translocates 9 protons, and produces 3 ATP, per rotation. The number of protons depends on the number of c subunits in the Fo c-ring, and it is now known that this is 10 in yeast Fo[33] and 8 for vertebrates[34]. Including one H+ for the transport reactions, this means that synthesis of one ATP requires 1+10/3=4.33 protons in yeast and 1+8/3 = 3.67 in vertebrates. This would imply that in human mitochondria the 10 protons from oxidizing NADH would produce 2.72 ATP (instead of 2.5) and the 6 protons from oxidizing succinate or ubiquinol would produce 1.64 ATP (instead of 1.5). This is well within the margin of experimental error described in a recent review[35].
blah blah blah [36].
.
Use of capital letters to designate the type of heme[edit]
Note Palmer's IUPAC article on nomencllature of electron transfer proteins makes no mention. Started by Caughey?)() who named his preparation of heme "heme A" to distinguish it from heme a, heme a', heme a* etc which were various modified forms of heme. In a later paper() he used capital letters for hemesA, B, and C. The practice was formalized by Wikstrom () who explained the usage: --- lower case when it is really a cytochrome, i.e. in a particular environment in a protein as opposed to the substance. For example cytochome oxidase (complex III) contains heme a and a3, for a total of 2 mol heme A/mol protein. Complex III contains cytochrome b with heme bH and bL, and cytochrome c1, for a total of 2 mol heme B and 1 mol heme C per mole of the complex. Papers by halestrap, berry and Rich follow this practice
Although not sanctioned/proscribed by official convention (e.g. Palmer)International Union of Biochemistry (1979) Enzyme Nomenclature(Academic, New York), pp. 593-601., the practice of designating hemes with upper case letters has been used by a number of authors (caughey, Halestrap, Wikstrom, rich, berry).
The practice of designating hemes with upper case letters was formalized in a footnote in a paper by Puustinen & Wikstrom [37] which explains under which conditions a capital letter should be used: "we prefer the use of capital letters to describe the heme structure as isolated. Lowercase letters may then be freely used for cytochromes and enzymes, as well as to describe individual protein-bound heme groups (for example, cytochrome bc, and aa3 complexes, cytochrome b5, heme c1 of the bc1 complex, heme a3 of the aa3 complex, etc)." In other words the chemical compound would be designated with a capital letter, but specific instances in structures with lowercase. Thus cytochrome oxidase, which has two A hemes (heme a and heme a31) in its structure, contains two moles of heme A per mole protein. Cytochrome bc1, with hemes bH, bL, and c1; contains heme B and heme C in a 2:1 ratio. The practice seems to have originated in a paper by Caughey and York in which the product of a new isolation procedure for the heme of cytochrome aa3 was designated heme A to differentiate it from previous preparations: "Our product is not identical in all respects with the heme a obtained in solution by other workers by the reduction of the hemin a as isolated previously (2). For this reason, we shall designate our product heme A until the apparent differences can be rationalized."[38]. In a later paper [39], Caughey's group uses capital letters for isolated heme B and C as well as A.
Biochemical process of fermentation of sucrose[edit]

The chemical equations below summarize the fermentation of sucrose (C12H22O11) into ethanol (C2H5OH). Alcoholic fermentation converts one mole of glucose into two moles of ethanol and two moles of carbon dioxide, producing two moles of ATP in the process.
The overall chemical formula for alcoholic fermentation is:
- C6H12O6 → 2 C2H5OH + 2 CO2
Sucrose is a dimer of glucose and fructose molecules. In the first step of alcoholic fermentation, the enzyme invertase cleaves the glycosidic linkage between the glucose and fructose molecules.
- C12H22O11 + H2O + invertase → 2 C6H12O6
Next, each glucose molecule is broken down into two pyruvate molecules in a process known as glycolysis.[40] Glycolysis is summarized by the equation:
- C6H12O6 + 2 ADP + 2 Pi + 2 NAD+ → 2 CH3COCOO− + 2 ATP + 2 NADH + 2 H2O + 2 H+
The chemical formula of pyruvate is CH3COCOO−. Pi stands for the inorganic phosphate.
The enzyme in step 3 is pyruvate decarboxylase.
Finally, pyruvate is converted to ethanol and CO2 in two steps, regenerating oxidized NAD+ needed for glycolysis:
- 1. CH3COCOO− + H+ → CH3CHO + CO2
catalyzed by pyruvate decarboxylase
- 2. CH3CHO + NADH+H+ → C2H5OH + NAD+
This reaction is catalyzed by alcohol dehydrogenase (ADH1 in baker's yeast)[41]
As shown by the reaction equation, glycolysis causes the reduction of two molecules of NAD+ to NADH. Two ADP molecules are also converted to two ATP and two water molecules via substrate-level phosphorylation.
Related processes[edit]
Fermentation of sugar to ethanol and CO2 can also be done by Zymomonas mobilis, however the path is slightly different since formation of pyruvate does not happen by glycolysis but instead by the Entner–Doudoroff pathway. Other microorganisms can produce ethanol from sugars by fermentation but often only as a side product. Examples are[42]
- Heterolactic acid fermentation in which Leuconostoc bacterias produce Lactate + Ethanol + CO2
- Mixed acid fermentation where Escherichia produce Ethanol mixed with Lactate, Acetate, Succinate, Formate, CO2 and H2
- 2,3-butanediol fermentation by Enterobacter producing Ethanol, Butanediol, Lactate, Formate, CO2 and H2
PMID reference[edit]
The term Fo stands for ologomycin sensitivity conferring factor (or fraction)[43].or [44] or maybe [45]
use this:
[46] or Use this [47]. It seems that Title is the only thing that is required (as long as PMID is provided?) [48]. Another template:this.[49]
and later this [51]
Complex II[edit]
Succinate dehydrogenase was first isolated as a two subunit enzyme (EC 1.3.99.1) lacking the two membrane subunits, called succinic dehydrogenase [52] [53].
The 4-subunit succinate:ubiquinone oxidorectase (EC 1.3.5.1) was first isolated by Hatefi and co-workers in 1962 and named "Complex II"[54] to differentiate it from the other three respiratory complexes described in that paper. Unrelated reference [55]. Which was further shown in a later paper,[56] and a follow-up work.[57]
- ^ Kyoko Shinzawa-ltoh; Hidefumi Ueda; Shinya Yoshikawa; Hiroshi Aoyama; Eiki Yamashita; Tomitake Tsukihara (1995). "Effects of Ethyleneglycol Chain Length of Dodecyl Polyethyleneglycol Monoether on the Crystallization of Bovine Heart Cytochrome c Oxidase". J. Mol. Biol. 246 (5): 572–575. doi:10.1016/S0022-2836(05)80106-8. PMID 7877177.
{{cite journal}}: Unknown parameter|last-author-amp=ignored (|name-list-style=suggested) (help) - ^ Pentaethylene glycol monododecyl ether at Sigma-Aldrich
- ^ "Potentiometric Studies Of The Secondary Phosphate Ionizations Of Amp, Adp, And Atp, And Calculations Of Thermodynamic Data For The Hydrolysis Reactions". Biochemistry. 1963. PMID 14069537.
- ^ "What are TeX, LaTeX and friends?".
- ^ Alexia Gaudeul (March 27, 2006). "Do Open Source Developers Respond to Competition?: The (La)TeX Case Study". SSRN 908946.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Leslie Lamport (April 23, 2007). "The Writings of Leslie Lamport: LaTeX: A Document Preparation System". Leslie Lamport's Home Page. Retrieved 2007-04-27.
- ^ A B Taylor, B S Smith, S Kitada, K Kojima, H Miyaura, Z Otwinowski, A Ito, J Deisenhofer (2001). "Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences". Structure. 9: 615–625. doi:10.1016/s0969-2126(01)00621-9. PMID 11470436.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hassan Sharifiyazdi, Fatemah Namazi, Ahmad Oryan, Reza Shahriari, Mostafa Razavi (2012). "Point mutations in the Theileria annulata cytochrome b gene is associated with buparvaquone treatment failure". Veterinary Parasitology. 187: 431–435. PMID 22305656.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Saitoh, I., Miyoshi, H., Shimizu, R., and Iwamura, H. (1992). "Comparison of structure of quinone redox site in the mitochondrial cytochrome-bc1 complex and photosystem II (QB site)". Eur. J. Biochem. 209: 73–79. doi:10.1111/j.1432-1033.1992.tb17262.x. PMID 1327783.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ K S Oyedotun, B D Lemire (1997). "The Carboxyl Terminus of the Saccharomyces cerevisiae Succinate Dehydrogenase Membrane Subunit, SDH4p, Is Necessary for Ubiquinone Reduction and Enzyme Stability*". J Biol Chem. 272: 31382–8. doi:10.1074/jbc.272.50.31382. PMID 9395469.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^
{{cite journal}}: Empty citation (help) - ^ Sebald W, Weiss H, and Jackl G (1972). "Inhibition of the Assembly of Cytochrome Oxidase in Neurospora crussa by Chloramphenicol". Eur. J. Biochem. 30: 413–417. PMID 4344826.
- ^ Romana Fato, Christian Bergamini, Marco Bortolus, Anna Lisa Maniero, Serena Leoni, Tomoko Ohnishi, and Giorgio Lenaz (2009). "Differential effects of mitochondrial Complex I inhibitors on production of reactive oxygen species". Biochim Biophys Acta. 1787: 384–392. doi:10.1016/j.bbabio.2008.11.003. PMC 2724837. PMID 19059197.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ^ von Jagow G, Ohnishi T. (1985). "The chromone inhibitor stigmatellin--binding to the ubiquinol oxidation center at the C-side of the mitochondrial membrane". FEBS Lett. 185: 311–5. doi:10.1016/0014-5793(85)80929-7. PMID 2987042.
{{cite journal}}: Vancouver style error: punctuation in name 2 (help) - ^ Trumpower BL, Edwards CA. (1979). "Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate:cytochrome c reductase complex of bovine heart mitochondria". J Biol Chem. 254: 8697–706. PMID 224062.
{{cite journal}}: Vancouver style error: punctuation in name 2 (help) - ^
{{cite journal}}: Empty citation (help) - ^ Hördt A, López MG, Meier-Kolthoff JP, Schleuning M, Weinhold L-M, Tindall BL, Gronow S, Kyrpides NC, Woyke T, Göker M. "Analysis of 1,000+ Type-Strain Genomes Substantially Improves Taxonomic Classification of Alphaproteobacteria". Frontiers in Microbiology. 11: 468. doi:10.3389/fmicb.2020.00468.
{{cite journal}}: Vancouver style error: name in name 5 (help)CS1 maint: unflagged free DOI (link) - ^ Thibodeau SN, Palmiter RD, Walsh KA. (1978). "Precursor of egg white ovomucoid. Amino acid sequence of an NH2-terminal extension". J Biol Chem. 253: 9018–23. PMID 721826.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Maynes, J.T., Cherney, M.M., Qasim, M.A., Laskowski Jr, M., James, M.N. (2005). "Structure of the subtilisin Carlsberg-OMTKY3 complex reveals two different ovomucoid conformations". Acta Crystallogr D Biol Crystallogr. 61: 580–588. PMID 15858268.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^
{{cite journal}}: Empty citation (help) - ^ a b JPEG. "Joint Photographic Experts Group, JPEG2000". Retrieved 2009-11-01.
- ^ "JAR File Specification". Oracle Corporation. Retrieved 2020-09-14.
- ^ a b Gosling, James; Joy, Bill; Steele, Guy; Bracha, Gilad. "The Java Language Specification, 2nd Edition". Archived from the original on August 5, 2011. Retrieved February 8, 2008.
- ^ ISO/IEC JTC 1/SC 29 (2009-05-07). "ISO/IEC JTC 1/SC 29/WG 1 - Coding of Still Pictures (SC 29/WG 1 Structure)". Archived from the original on 2013-12-31. Retrieved 2009-11-11.
{{cite web}}: CS1 maint: numeric names: authors list (link) - ^ ISO/IEC JTC 1/SC 29. "Programme of Work, (Allocated to SC 29/WG 1)". Archived from the original on 2013-12-31. Retrieved 2009-11-07.
{{cite web}}: CS1 maint: numeric names: authors list (link) - ^ ISO. "JTC 1/SC 29 - Coding of audio, picture, multimedia and hypermedia information". Retrieved 2009-11-11.
- ^ "JSR 56: Java Network Launching Protocol and API". Retrieved 2020-09-14.
- ^ a b c "T.81 – DIGITAL COMPRESSION AND CODING OF CONTINUOUS-TONE STILL IMAGES – REQUIREMENTS AND GUIDELINES" (PDF). CCITT. September 1992. Retrieved 12 July 2019.
- ^ "ECMAScript 2020 Language Specification".
- ^ "Standard ECMA-404 - The JSON Data Interchange Syntax" (PDF). Ecma International. December 2017. p. 1, footnote. Retrieved 27 October 2019.
- ^ "ISO/IEC 21778:2017". ISO. Retrieved 29 July 2019.
- ^ "JSR 245: JavaServer Pages 2.1". jcp.org. 2013-06-12. Retrieved 2020-09-14.
- ^ Stock D, Leslie AG, Walker JE (1999). "Molecular architecture of the rotary motor in ATP synthase". Science. 286: 1700–5. PMID 10576729.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Watt, I.N., Montgomery, M.G., Runswick, M.J., Leslie, A.G.W., Walker, J.E. (2010). "Bioenergetic Cost of Making an Adenosine Triphosphate Molecule in Animal Mitochondria". Proc.Natl.Acad.Sci.USA. 107: 16823. PMID 20847295.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ P.Hinkle (2005). "P/O ratios of mitochondrial oxidative phosphorylation". Biochimica et Biophysica Acta. 1706: 1–11. PMID 15620362.
- ^ Henderson R1, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. (1990). "Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy". J Mol Biol. 213: 899–929. PMID 2359127.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ Puustinen A, Wikström M. (1991). "The heme groups of cytochrome o from Escherichia coli". Proc Natl Acad Sci U S A. 88: 6122–6. PMID 2068092.
- ^ CAUGHEY WS, YORK JL. (1962). "Isolation and some properties of the green heme of cytochrome oxidase from beef heart muscle". J Biol Chem. 237: 2414–6. PMID 13877421.
- ^ Caughey WS, Smythe GA, O'Keeffe DH, Maskasky JE, Smith MI (1975). "Heme A of cytochrome c oxidase. Structure and properties: comparisons with hemes B, C, and S and derivatives". J Biol Chem. 250: 7602–22. PMID 170266.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Stryer, Lubert (1975). Biochemistry. W. H. Freeman and Company. ISBN 0-7167-0174-X.
- ^ Raj SB, Ramaswamy S, Plapp BV. "Yeast alcohol dehydrogenase structure and catalysis". Biochemistry. 53: 5791-803. doi:10.1021/bi5006442. PMID 25157460.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ http://web.oranim.ac.il/courses/microbiology/Bacterial%20Fermentation%20Nature.pdf
- ^ PMID=4223640
- ^ Kagawa Y, Racker E. (1966). "Partial resolution of the enzymes catalyzing oxidative phosphorylation. 8. Properties of a factor conferring oligomycin sensitivity on mitochondrial adenosine triphosphatase". Journal of Biological Chemistry. 241: 2461–2466.
- ^
{{cite journal}}: Empty citation (help) - ^ ??? (???). "???". ???. ???: ??-??. PMID ???.
{{cite journal}}:|author=has numeric name (help); Check|pmid=value (help); Check date values in:|year=(help) - ^ "this is the title". 1951. PMID 4223640.
{{cite journal}}: Cite journal requires|journal=(help) - ^ "this is the title". PMID 4223640.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Venable, W. H.; Eckerle, K. L. "Didymium Glass Filters for Calibrating the Wavelength Scale of Spectrophotometers SRMs 2009, 2010, 2013 and 2014".
- ^ "Bioenergetic Cost of Making an Adenosine Triphosphate Molecule in Animal Mitochondria". Proc.Natl.Acad.Sci.USA. 107: 16823. 2010. PMID 20847295.
{{cite journal}}: Cite uses deprecated parameter|authors=(help) - ^ "Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM". Elife. 4: e10180–e10180. 2015. PMID 26439008.
{{cite journal}}: Cite uses deprecated parameter|authors=(help) - ^ BERNATH P, KEARNEY EB, SINGER TP. (1956). "Studies on succinic dehydrogenase. II. Isolation and properties of the dehydrogenase from beef heart". J Biol Chem. 223: 599–613. PMID 13385208.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ WANG TY, TSOU CL, WANG YL (1958). "Studies on succinic dehydrogenase. II. Further observations on the properties of the enzyme and its prosthetic group". Sci Sin. 7: 65–74. PMID 13529046.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Y. Hatefi, A. G. Haavik, L. R. Fowler, and D. E. Griffiths (1962). "Studies on the Electron Transfer System: XLII. RECONSTITUTION OF THE ELECTRON TRANSFER SYSTEM". J. Biol. Chem. 1962 237:. 237: 2661–9. PMID 13905326.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Thierry Ferreira, A. Brett Mason and Carolyn W. Slayman (2001). "The Yeast Pma1 Proton Pump: a Model for Understanding the Biogenesis of Plasma Membrane Proteins". J Biol Chem. 276: 29613–29616. doi:10.1074/jbc.R100022200.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Beyenbach, KW; Wieczorek, H (Feb 2006). "The V-type H+ ATPase: molecular structure and function, physiological roles and regulation". J Exp Biol. 209 (4): 577–89. PMID 16449553.
- ^ Kim, MS; Jang, J; Ab Rahman, NB; Pethe, K; Berry, EA; Huang, LS (5 June 2015). "Isolation and Characterization of a Hybrid Respiratory Supercomplex Consisting of Mycobacterium tuberculosis Cytochrome bcc and Mycobacterium smegmatis Cytochrome aa3". The Journal of biological chemistry. 290 (23): 14350–60. PMID 25861988.


![{\displaystyle [R]+[L]{\underset {k_{\text{off}}}{\overset {k_{\text{on}}}{\rightleftharpoons }}}[RL]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/9125b7abb98ebb3371740699753f79d45558d2bf)
![{\displaystyle [R]+[L]{\underset {K_{\text{D}}}{\rightleftharpoons }}[RL]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/1875d4d9d666184bca79a8594b5c7bf601f19f16)
![{\displaystyle k_{\text{on}}[R][L]=k_{\text{off}}[RL]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/b3389e39bfe13d0e3f95b6ee07d3c4d6f7684ca4)
![{\displaystyle {\bar {n}}={\frac {[RL]}{[R]+[RL]}}={\frac {[L]}{K_{d}+[L]}}=(1-{\bar {n}}){\frac {[L]}{K_{d}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/0749e8b8d666bfa1a6625feb26628f5f7da37089)
![{\displaystyle Kd=[R]{\frac {[L]}{[RL]}}(3)}](https://wikimedia.org/api/rest_v1/media/math/render/svg/a8fef4c34e6126361b8d5172db5feb6167f07056)
![{\displaystyle Kd{\frac {[RL]}{[L]}}=[R]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/b5c83e2dd210c4ea6144bd9b3770193102c4eab4)
![{\displaystyle Kd{\frac {[RL]}{[L]}}=n-[RL]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/ecc4e0b1df0e4326503911b2492484f8068df4ff)
![{\displaystyle [RL]=n-Kd{\frac {[RL]}{[L]}}(4)}](https://wikimedia.org/api/rest_v1/media/math/render/svg/19a4a3c1997c415b11a74e86abd406dc977610a3)

